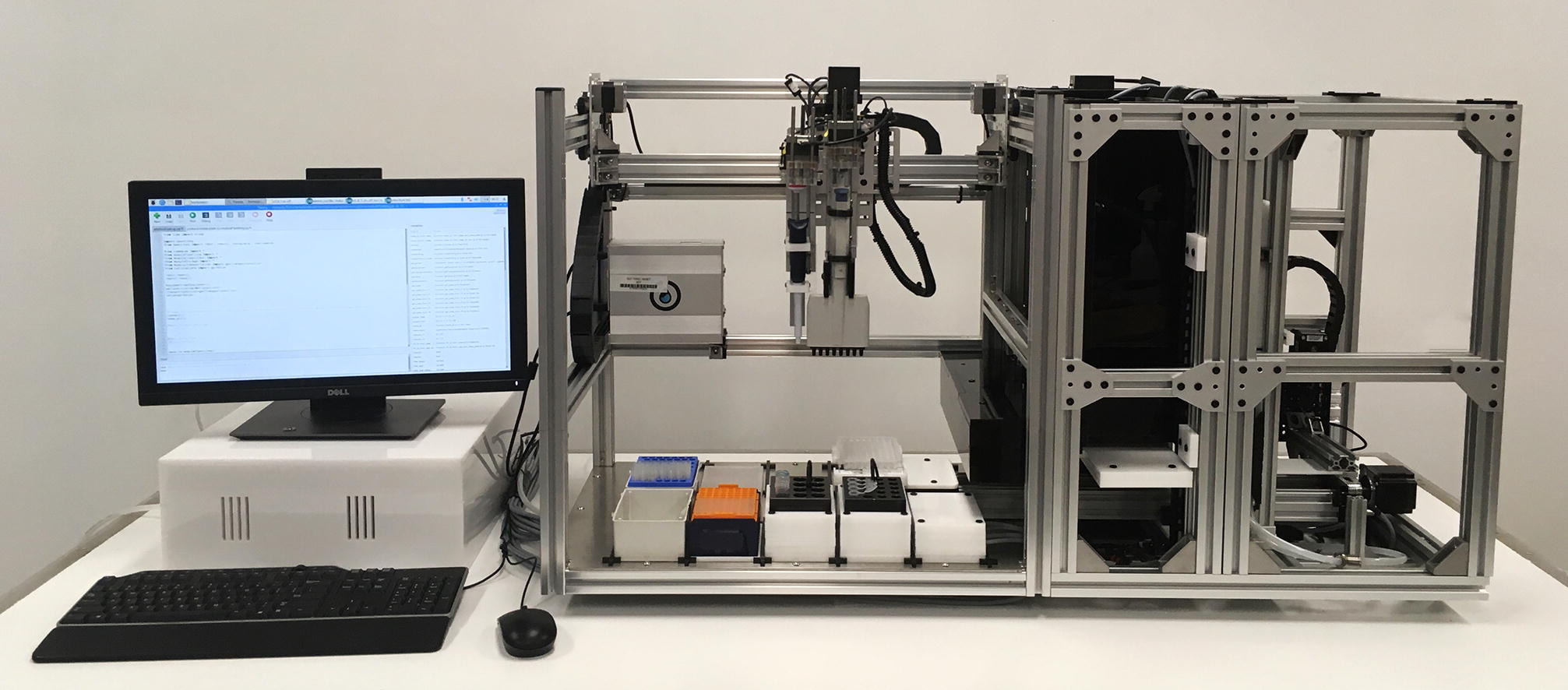
The OpenWorkstation is a cutting-edge modular open-source platform designed to enhance automation in in vitro workflows. By utilizing customizable modules, this workstation enables seamless integration of various functions for liquid handling, transportation, and crosslinking processes. This approach addresses the high costs and inflexibility of traditional laboratory automation systems, offering an affordable and efficient solution for academic and research institutions.
Key Features
- Modular Design: Allows researchers to customize their setup with various interchangeable modules tailored to specific laboratory needs.
- Cost-Effective: The base frame with a transportation module costs approximately $900, while additional modules range from $675 to $4,070, depending on functionality.
- Open Source: All design files and operational instructions are available for free, promoting community collaboration and innovation.
- Automation Capabilities: Automates repetitive tasks such as pipetting, transporting samples, and initiating photo-induced polymerization of hydrogels, improving reproducibility and efficiency.
- Scalable Functionality: Easily add or remove modules to adapt to changing experimental requirements, making it suitable for a wide range of applications in life sciences.
Specifications
| Specification | Details |
|---|---|
| Device Name | OpenWorkstation |
| Base Cost | $900 |
| Pipetting Module Cost | $4,070 |
| Crosslinker Module Cost | $675 |
| Storage Module Cost | $690 |
| Open Source License | Creative Commons Attribution-ShareAlike 4.0 (CC BY-SA 4.0) |
| Source File Repository | OpenWorkstation GitHub |
Build Instructions
- Assemble Modules: Build the base frame and attach the necessary modules according to the design specifications.
- Setup Control Box: Connect electronics and power supplies to manage the workstation’s operations.
- Install Software: Set up the Python API to facilitate control of the various modules and to run experimental protocols.
- Functional Testing: Conduct tests on each module to ensure they operate correctly and reliably.
Operation Instructions
- Prepare Samples: Load samples into the designated modules (e.g., pipetting or storage).
- Configure Protocol: Use the Python-based interface to define experimental parameters and protocols.
- Run Automation: Start the automated workflow, monitoring the progress through the interface.
Validation
The OpenWorkstation has been validated through case studies demonstrating its successful application in cell culture workflows and high-throughput studies, showcasing its effectiveness in producing reliable results in various biological experiments.
OpenWorkstation represents a significant advancement in laboratory automation, combining modular design with open-source principles to enhance the reproducibility and efficiency of in vitro experiments. Its affordability and adaptability make it an ideal solution for academic laboratories looking to innovate their workflows.