pip install alphapeptstats or download it from GitHub.
With the increasing use of mass spectrometry in proteomics, researchers need robust tools to interpret complex datasets. AlphaPeptStats, an open-source Python package developed by Elena Krismer, Isabell Bludau, Maximilian T. Strauss, and Matthias Mann, offers a scalable and automated solution for statistical analysis in mass spectrometry-based proteomics. This tool integrates seamlessly with popular proteomics workflows, providing functionalities like data normalization, imputation, and visualization, enabling researchers to gain valuable insights without extensive programming skills.
About AlphaPeptStats
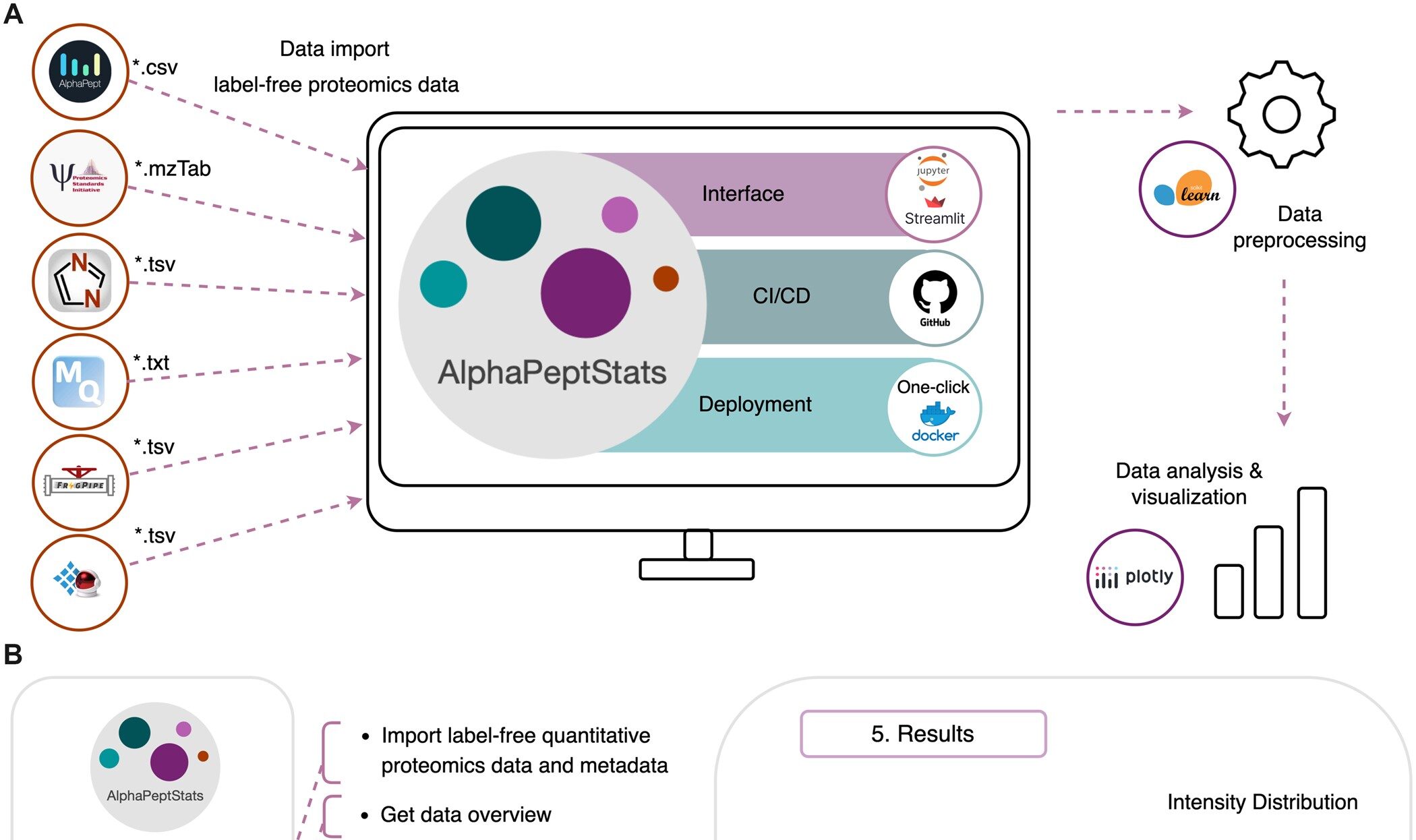
AlphaPeptStats is a comprehensive package designed to streamline and automate the analysis of label-free proteomics data. It offers robust statistical and visualization tools, allowing users to process, analyze, and interpret their data in a standardized way. Built within the AlphaPept framework, this package leverages popular Python libraries like NumPy, Pandas, SciPy, and Plotly, offering a modular structure for data exploration, from preprocessing through to visualization. With a simple installation process and a user-friendly interface, AlphaPeptStats ensures accessibility for both proteomics experts and non-experts alike.
Key Features
- Automated Statistical Analysis: Integrates statistical tests such as t-tests, ANOVA, and hierarchical clustering, all accessible through an easy-to-use Python interface.
- Data Visualization: Generates publication-ready visualizations, including volcano plots, heatmaps, and scatter plots.
- High Compatibility: Supports data formats from popular software like MaxQuant, DIA-NN, and FragPipe, allowing integration with diverse proteomics workflows.
- Easy Installation: Available on PyPI with one-click installers for Windows, macOS, and Linux, and Docker support for cloud deployment.
- Extensive Documentation and Tutorials: Provides Jupyter notebooks and interactive tutorials, helping users explore the package’s full capabilities with ease.
Design and Components
AlphaPeptStats is built on a range of community-tested scientific libraries, making it both reliable and flexible:
- Data Handling: AlphaPeptStats processes proteomics data through a Python class,
DataSet, enabling robust data manipulation and tracking of preprocessing steps. - Preprocessing Options: Allows contaminant removal, normalization, and imputation. It uses advanced techniques, like random forest imputation, to enhance data quality.
- Visualization and Analysis Tools: Equipped with functions for complex visualizations, such as principal component analysis (PCA) and volcano plots. It also includes GO term enrichment analysis, facilitating biological insights from statistical results.
Cost and Accessibility
AlphaPeptStats is freely available under the Apache License on GitHub, ensuring open access and adaptability. With a high testing coverage of 98%, it’s reliable and maintained to a professional standard. Its compatibility with Docker and Streamlit for GUI-based use makes it easy to deploy on local machines or in cloud environments.
Performance and Results
In testing, AlphaPeptStats demonstrated robust performance, especially in data imputation and differential expression analysis. Researchers applied AlphaPeptStats to proteomic datasets, where it reliably identified biologically significant patterns and improved reproducibility. For instance, in a study on non-alcoholic liver disease, the package facilitated data normalization and biomarker discovery, helping researchers uncover potential disease markers.
How to Use AlphaPeptStats
AlphaPeptStats is accessible for both coding and non-coding users:
- Installation: Install AlphaPeptStats from PyPI using
pip install alphapeptstats. - Data Loading: Import proteomics data from compatible software like MaxQuant or DIA-NN.
- Data Processing and Analysis: Use the built-in preprocessing and statistical functions to clean, normalize, and analyze your data.
- Visualization: Generate interactive plots for publication-ready figures or further data exploration.
- Optional GUI: Use the Streamlit-based GUI for a more accessible, browser-based experience.
Documentation and tutorials are available on GitHub to guide users through each step of the workflow.
AlphaPeptStats is a powerful, open-source tool for the proteomics community, enabling accessible, reproducible, and high-quality analysis of mass spectrometry data. With its modular structure, extensive documentation, and robust testing, AlphaPeptStats stands out as an essential tool for both routine and advanced proteomics analysis, making it invaluable for academic and clinical research settings.